Neuronal networks are the physical substrate of the information flow inside the brain. In LNMC we are interested in neuronal micro- and mesoscale connectivity, since this connectitvity defines the paths of information flow within and acros brain structures. In particular, we wish to classify, quantify and reconstruct the neuronal micro- and meso scale connectivity of the rodent brain with standardized neuroanatomical methods. To map the micro scale connectivity we use neurolucida to digitally reconstruct individual neurons in 3D. We presently have almost 1000 neuron reconstructions, which can be used to identify the physical constraints and most likely targets of local connectivity [1]. In combination with paired electrophysiological recordings this data is used to map local connectivity of cortical microcircuits. We are also currently developing new techniques for whole mount imaging, with the aim to map meso scale afferent and efferent projections of the primary somatosensory cortex.
Approach
Single cell Reconstruction
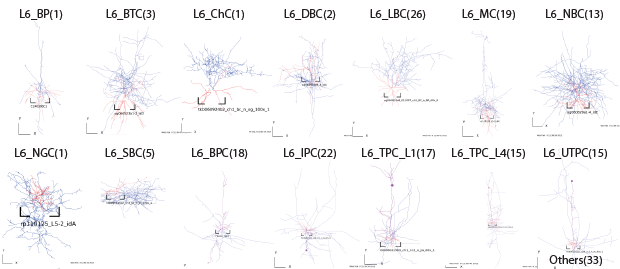
We routinely stain and reconstruct biocytin filled, electrophysiologically characterized cells in acute slice preparations. Continued effort in this direction has so far generated a database of approximately 1000 reconstructed unique cell morphologies. This resulting dataset forms the basis of all generative algorithms used by the Blue Brain Project to create unique virtual cells used in their cortical simulations. Together with gene expression data and electrophysiological characterization we hope to fully characterize neocortical cell populations
Layer1
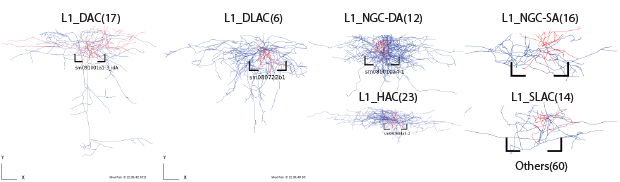
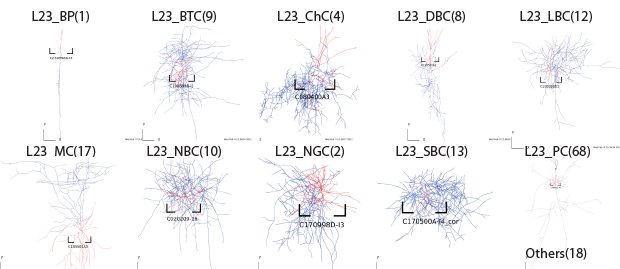
Layer2/3
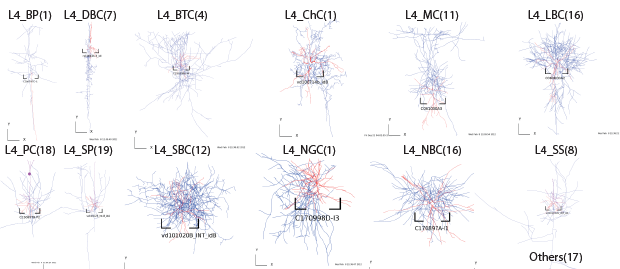
Layer4
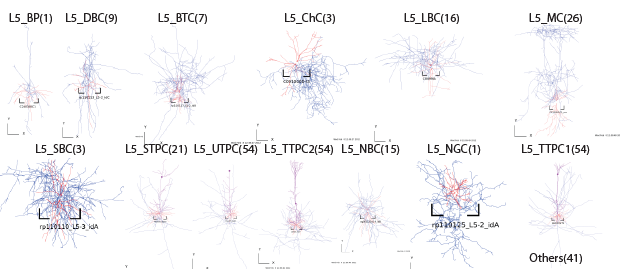
Layer5
Layer6
Immunohistochemical characterization of neuron subtypes
Different neuron subtypes express different combinations of genes and proteins. The expression of these protein markers can be visualized and used to identify different neuronal populations in brain slices. We routinely use immunohistochemistry (IHC), 3D confocal microscopy and stereological methods to calculate cell densities of different neuron populations in different regions of the brain. Combining IHC data with the single-cell gene expression data will allow us to map the anatomical distribution of neuronal populationes defined by their gene expression profiles.